ideogram
Version:
Chromosome visualization for the web
72 lines (51 loc) • 4.39 kB
Markdown
# ideogram
[](https://app.travis-ci.com/github/eweitz/ideogram)
[](https://coveralls.io/github/eweitz/ideogram)
[Ideogram.js](https://eweitz.github.io/ideogram/) is a JavaScript library for [chromosome visualization](https://speakerdeck.com/eweitz/designing-genome-visualizations-with-ideogramjs).
Ideogram supports drawing and animating genome-wide datasets for [human](https://eweitz.github.io/ideogram/human), [mouse](https://eweitz.github.io/ideogram/mouse), and [many other eukaryotes](https://eweitz.github.io/ideogram/eukaryotes). The [Ideogram API](https://github.com/eweitz/ideogram/blob/master/api.md) for annotations supports [histograms](https://eweitz.github.io/ideogram/annotations-histogram), [heatmaps](https://eweitz.github.io/ideogram/annotations-heatmap), [overlays](https://eweitz.github.io/ideogram/annotations-overlaid), and points of arbitrary shape and color layered in [tracks](https://eweitz.github.io/ideogram/annotations-tracks). Ideogram can depict haploid, [diploid](https://eweitz.github.io/ideogram/ploidy-basic) or higher ploidy genomes (e.g. plants), as well as aneuploidy, [genetic recombination](https://eweitz.github.io/ideogram/ploidy-recombination), and [homologous features](https://eweitz.github.io/ideogram/homology-basic) between chromosomes.
Ideogram can be embedded as a [reusable component](https://github.com/eweitz/ideogram#usage) in any web page or application, and leverages D3.js and SVG to achieve fast, crisp client-side rendering. You can also integrate Ideogram with JavaScript frameworks like [Angular](https://github.com/eweitz/ideogram/tree/master/examples/angular), [React](https://github.com/eweitz/ideogram/tree/master/examples/react), and [Vue](https://github.com/eweitz/ideogram/tree/master/examples/vue), as well as data science platforms like [R](https://github.com/eweitz/ideogram/tree/master/examples/r) and [Jupyter Notebook](https://github.com/eweitz/ideogram/tree/master/examples/jupyter).
[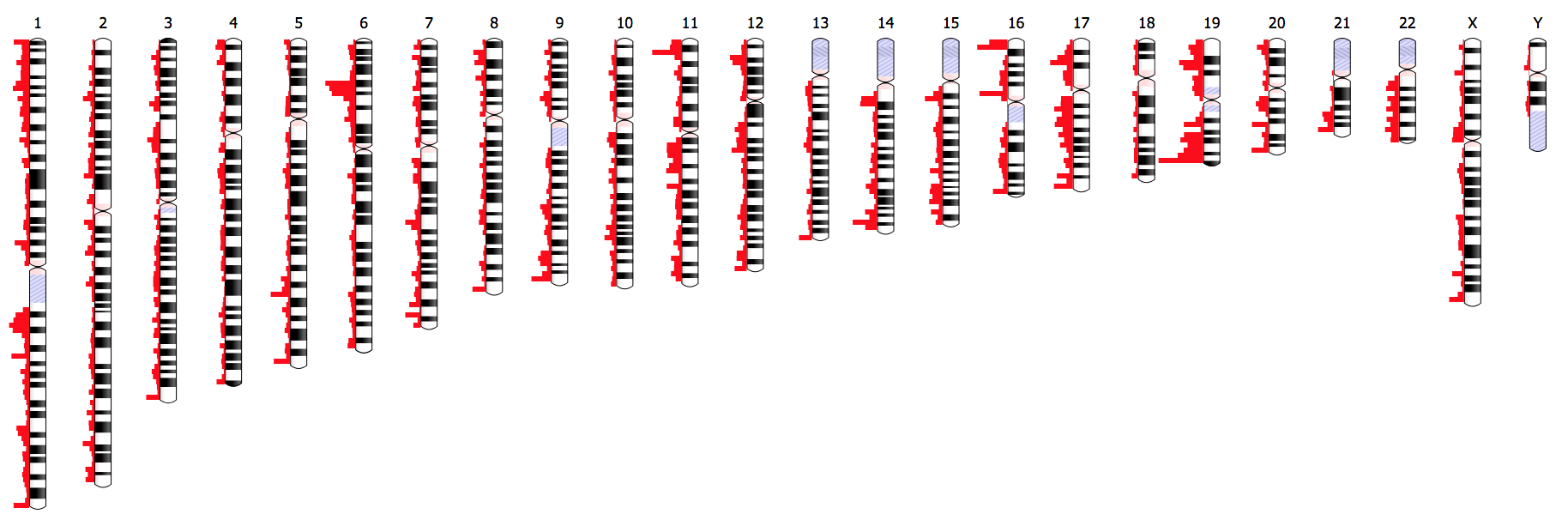](https://eweitz.github.io/ideogram/annotations_histogram.html)
Check out [live examples](https://eweitz.github.io/ideogram/), get [up and running](#installation) with your own deployment, skim [basic usage](#usage), or dive into the [full API](api.md)!
A [broader overview](https://speakerdeck.com/eweitz/ideogramjs-chromosome-visualization-with-javascript) is also available. It discusses Ideogram's chromosome biology, technical architecture, and more.
# Installation
To link directly to the latest release, copy this snippet:
```
<script src="https://cdn.jsdelivr.net/npm/ideogram@1.37.0/dist/js/ideogram.min.js"></script>
```
You can also easily use the library locally:
```
$ cd <your local web server document root>
$ git clone https://github.com/eweitz/ideogram.git
```
Then go to [http://localhost/ideogram/examples/](http://localhost/ideogram/examples/).
Or, if you use npm:
```
npm install ideogram
```
You can then [import](https://developer.mozilla.org/en-US/docs/Web/JavaScript/Reference/Statements/import) Ideogram into an application like so:
```
import Ideogram from 'ideogram';
```
# Usage
```html
<head>
<script src="https://cdn.jsdelivr.net/npm/ideogram@1.37.0/dist/js/ideogram.min.js"></script>
</head>
<body>
<script>
var ideogram = new Ideogram({
organism: 'human',
annotations: [{
name: 'BRCA1',
chr: '17',
start: 43044294,
stop: 43125482
}]
});
</script>
</body>
```
Many more usage examples are available at https://eweitz.github.io/ideogram/.
You can also find examples of integrating Ideogram with JavaScript frameworks like [Angular](https://github.com/eweitz/ideogram/tree/master/examples/angular), [React](https://github.com/eweitz/ideogram/tree/master/examples/react), and [Vue](https://github.com/eweitz/ideogram/tree/master/examples/vue), as well as data science platforms like [R](https://github.com/eweitz/ideogram/tree/master/examples/r) and [Jupyter Notebook](https://github.com/eweitz/ideogram/tree/master/examples/jupyter).
# API
See the [Ideogram API reference](api.md) for detailed documentation on configuration options and methods.